Excel-SBOL Converter: Creating SBOL from
Excel Templates and Vice Versa
Jeanet Mante,
†
Julian Abam,
†
Sai P. Samineni,
†
Isabel M. P¨otzsch,
‡
Prubhtej
Singh,
¶
Jacob Beal,
§
and Chris J. Myers
∗,†
†University of Colorado Boulder
‡University of Cambridge
¶Guru Gobind Singh Indraprastha University
§Raytheon BBN Technologies
E-mail: chris.myers@colorado.edu
Abstract
Standards support synthetic biology research by enabling the exchange of compo-
nent information. However, using formal representations, such as the Synthetic Biology
Open Language (SBOL), typically requires either a thorough understanding of these
standards or a suite of tools developed in concurrence with the ontologies. Since these
tools may be a barrier for use by many practitioners, the Excel-SBOL Converter was
developed to allow easier use of SBOL and integration into existing workflows. The
converter consists of two Python libraries: one that converts Excel templates to SBOL,
and another that converts SBOL to an Excel workbook. Both libraries can be used
either directly or via a SynBioHub plugin. We illustrate the operation of the Excel-
SBOL Converter with two case studies: uploading experimental data with the study’s
metadata linked to the measurements and downloading the Cello part repository.
1
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint
Keywords
Excel, SBOL, conversion, Excel-to-SBOL, SBOL-to-Excel, ontologies
Introduction
Synthetic biology is bringing together engineers and biologists to design biological circuits
for a variety of applications in energy, medicine, and bio-manufacturing.
1
Associated with
this interdisciplinary movement is the need for tools that support reusability and supple-
ment the current understanding of genetic sequences. To satisfy this need, synthetic biology
communities across the world have developed tools and ontologies to help describe their
unique semantic annotations.
2–15
Shared representations for data and metadata, grounded
in well-defined ontology terms, can help reduce confusion when sharing materials between
researchers.
16
The Synthetic Biology Open Language (SBOL)
6
has been developed to ad-
dress this challenge. SBOL provides a standardized format for the electronic exchange of
information on the structural and functional aspects of biological designs, supporting the use
of engineering principles such as abstraction, modularity, and standardization for synthetic
biology. Many tools have been created that work with SBOL,
17–26
including the SynBioHub
repository for storing and sharing designs.
27
The original SBOL has been developed further
to allow other data types to be represented and more information to be captured. This has
lead to SBOL2
28
and SBOL3.
29
Using formal representations such as SBOL, however, typically requires either a thorough
understanding of these standards or a suite of tools developed in concurrence with the
ontologies.
30
Unfortunately, this poses a significant barrier for scientists not trained to work
with such abstractions. Not using standards makes it difficult to share and reuse parts.
31–36
Therefore, the time and effort required to find parts is much greater, and the ability to use
the tools to automate design is limited.
One approach to lowering the barrier for use of ontologies was demonstrated by the Sys-
2
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint
tems Biology for Micro-Organisms (SysMO) consortium.
37
In SysMO, the MicroArray Gene
Expression Markup Language (Mage-ML) was set up as an XML schema,
38
and users were
expected to submit data to the SysMO Assets Catalogue (called SEEK) in XML format in
order to publish work. To allow the use of the Mage-ML language without having to un-
derstand XML, the RightField tool was created.
39
This tool is an ontology annotation and
information management application that can add constrained ontology term selection to Ex-
cel spreadsheets. It enables administrators to create templates with controlled vocabularies,
such that the scientists utilizing the tool would never actually see the raw RightField, only
the more familiar Excel spreadsheet interface. Spreadsheets are a popular interface as many
biological workflows already use spreadsheets and comma separated values (CSV) files. Fur-
thermore, several popular tools use spreadsheets and CSV files as inputs or outputs, including
Addgene (https://www.addgene.org/), and Opentrons (https://opentrons.com/).
Users of SBOL and SynBioHub have also faced a steep learning curve for understanding
the underlying ontology: as assessed in,
40
“For successful use and interpretation of metadata
presented in SynBioHub, the semantic annotation process should be biologist-friendly and
hide the underlying RDF predicates.” Recently, SynBio2Easy was published as a command
line tool to convert Excel spreadsheets of plasmids to SBOL.
41
The tool was designed to
enable several steps of a specific workflow for designing and depositing Synechocystis plasmids
into a public SynBioHub repository.
Embracing the same spreadsheet-based interface as these prior works, this paper presents
the Excel-SBOL Converter, a tool designed to provide a simple way for users to generate
and visualize SBOL data without needing a detailed understanding of the underlying on-
tology and associated technologies. Unlike SynBio2Easy, our converter generalizes beyond
plasmids to multiple kinds of SBOL data, as well as allowing customization of the tem-
plates. This converter thus provides a simple way for users to manage data by allowing users
to both download SBOL into Excel templates and to submit Excel templates for conver-
sion into SBOL. This paper presents the architecture and key engineering decisions for the
3
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint

Excel-SBOL Converter, as well as two case studies that illustrate its operation: uploading
experimental data and downloading a repository of parts.
Results
The Excel-SBOL Converter enables researchers that are more comfortable with Excel spread-
sheets to make use of SBOL repositories and tools without having to manipulate or under-
stand the SBOL data standard. The Excel-SBOL Converter is currently implemented as two
separate libraries: the Excel-to-SBOL library is used to convert Excel spreadsheets format-
ted using pre-designed templates into SBOL data, while the SBOL-to-Excel library is used
to convert SBOL data into Excel spreadsheets. These two libraries taken together enable
data to be converted between Excel spreadsheets and SBOL data as shown in Figure 1.
Figure 1: The Excel-SBOL Converter consists of two libraries: 1) Excel-to-SBOL allows the
use of Excel templates to create SBOL data, and 2) SBOL-to-Excel allows SBOL data to
be converted to Excel spreadsheets. Using the two libraries together allows the creation,
uploading, downloading, editing, and re-uploading of SBOL data. Additionally, it allows
collaboration between users who prefer spreadsheets and those using software tools that
require data to be provided in SBOL format.
4
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint

Excel-to-SBOL
The Excel-to-SBOL library converts Excel spreadsheet templates into SBOL data. While ini-
tially designed based on fixed spreadsheet templates developed in the Defense Advanced Re-
search Projects Agency (DARPA) Synergistic Discovery and Design (SD2) program (https:
//sd2e.org/)), the library has since been generalized to allow more flexibility. With this
approach, a user needs to have only minimal knowledge of SBOL, while a template designer
needs knowledge of both the SBOL data representation and the Excel-to-SBOL template
structure.
Excel Spreadsheet Templates
As an example, let us consider an Excel spreadsheet template that can organize genetic parts
into several part collection libraries. This example is composed of one Excel File containing
2 data sheets:
1. A sheet describing the part collection libraries (Figure 2).
2. A sheet describing the genetic parts (Figure 3).
Figure 2: Collections sheet. This is where the users add collections and a description of the
collection.
In order to use this template, a user would enter the library names and their descriptions
into the collections sheet. The user would then enter each genetic part name, the collection it
is a part of, any sequence alterations, part description, data source prefix (PubMed, GenBank
etc), Data Source (e.g. the PubMed Id), source organism, whether the part is circular, and
the sequence (the final column is automatically generated).
5
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint

Figure 3: Parts sheet. This is where the users add parts and a description of the parts.
Additionally, parts can be added to collections specified in the collections sheet (Figure 2)
by adding collection names to the “Collection” column.
Excel Spreadsheet Template Programming
Excel-to-SBOL is designed to parse a variety of interlinking SBOL object types in a flexible
manner, whilst maintaining code simplicity. This is achieved via two main innovations: an
initialization sheet, which lists all sheets to be processed, and a column definitions sheet,
which provides conversion parameters for each object property.
The initialization sheet is a list of all sheets to be processed (Figure 4). The initialization
sheet enables the conversion of multiple sheets. The first column in the initialization sheet is
the sheet name, while the rest indicate how it is to be processed. Specifically, these columns
indicate whether a conversion should be performed (if True it contains SBOL objects, if
False it is ignored except to potentially convert cell values), which row sheet data starts on,
whether collection metadata is present (and if so, the number of rows and which columns
it can be found in), and whether a collection description is present (and if so, where it can
be found). Additionally, further columns may be added to this sheet which are applied to
all objects it references. For example, in Figure 4 Molecule Type has been added and filled
out for the library sheet. This indicates that all parts on the library sheet have the molecule
type DNA.
The column definitions sheet is used for three things: conversion of cell values to a ma-
chine readable format, format checking of the cells, and conversion of cell values to SBOL.
The column definition sheet for our example is shown in Figure 5. The first two entries
identify the sheet and column that conversion is being defined for. The SBOL Term field
indicates the property that is found in this location, and how it should be converted into
6
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint

Figure 4: Example initialization sheet. The left most column contains the names of the
sheets to be processed. Next the ‘Convert’ column indicates whether the sheet contains
SBOL objects (contains SBOL objects if TRUE). The ‘Lib Start Row’ column indicates on
which zero-indexed row the information on the sheet starts. The next columns indicate if
collection metadata and description are provided or not, and if so, where they can be found.
All columns after Descript Cols are columns that contain information that is the same across
all items in the sheet.
SBOL or a custom data object. This definition begins with a short prefix for the names-
pace (e.g., “sbol”) followed by an underscore and the property name itself. Most terms
will lead to a simple addition or the property, however some (e.g. sbol sequence) call spe-
cialized functions that do more than just add a property (e.g. also creates a sequence
object). Additional fields indicate the namespace URL (Uniform Resource Locator) to use
for the property, the type of the value (e.g., String or URI (Uniform Resource Identifier)),
whether the property can have multiple values and the character(s) on which the field is
split for conversion into a list of values. Additional value checking can be performed with
a “pattern” field that provides a series of regular expressions that the property value is
to be checked against, separated by quotations. For example: “ ∧ [a − zA − Z\s ∗] + $”
“https : \/\/www.ncbi.nlm.nih.gov/nuccore/. ∗” is used for checking sequence entries, indi-
cating that entries should either contain only alphabetical characters and spaces or be URLs
starting with https://www.ncbi.nlm.nih.gov/nuccore/.
The remaining fields are used to define mechanisms that can be used for conversion from
human readable to machine readable forms. The first option is to use the Tyto software to
convert a string to an ontology term.
42
In this case, the ontology to look the terms up in
must also be given (“Ontology Name”). For example, in Figure 4, Role (on the Init Sheet)
is converted using the sequence Ontology (SO).
43
7
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint

Figure 5: Example column definitions sheet. The Sheet Name and Column Name entries
identify the sheet and column within the sheet that is being defined. Next, the SBOL Term
indicates the encoding property for the cell. The Namespace URL is the namespace to be
used with the encoding property, and the Type column indicates the type of data expected
(i.e. URI or String). The Split On column is used to split a single column into multiple
values for a property. The Pattern column contains regular expressions to check the cell
values after they have undergone conversion. The following columns contain conversion
information to go from human to machine-readable values. Tyto Lookup uses the Tyto
42
library to perform lookups according to the “Ontology Name” Column. The library performs
lookups according to the “Ontology Name” Column. Sheet lookup takes the cell value and
converts it to another value like a lookup dictionary. The dictionary is created based on the
“Lookup Sheet Name”, “From Col”, and “To Col”. Replacement Lookup is a special case of
sheet lookup. In this case, the cell value is expected to be prefix:value. Object ID Lookup
converts the cell value to an SBOL Object URI. The “Parent Lookup” column indicates the
directionality of the object reference.
8
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint

The second option is to lookup the ontology term using another sheet within the spread-
sheet as a dictionary (see for example Figure 6). When this option is selected, the user must
also specify the sheet to use for the lookup (“Lookup Sheet Name”), the column in which
the human readable value is found (“From Col”), and the column in which the machine
readable ontology term is found (“To Col”). For example, following the dictionary shown
in Figure 6), TRUE in the Circular column is converted to http://purl.obolibrary.org/
obo/SO_0000988 (the Sequence Ontology Role for circular) and false means no additional
role is added.
Figure 6: Lookup Sheet (circular types). This spreadsheet is used in a Lookup replacement
and converts human readable terms to machine readable ones. In this case the value TRUE
in the circular column leads to the Sequence Ontology term for circular whilst FALSE leads
to no addtion.
Replacement Lookup is a special case of sheet lookup. In this case, the cell value is
expected to be prefix:value, e.g. PMID:24295448, representing the data source is a PubMed
id with the value 24295448. Here the lookup works by using the prefix as the key and pulls a
value from the “To Col” and inserts the cell suffix in the place of {REPLACE HERE}
(Figure 7). So for the case PMID:24295448, the PMID key gives the string https://
pubmed.ncbi.nlm.nih.gov/{REPLACE_HERE}, which is changed to https://pubmed.ncbi.
nlm.nih.gov/24295448. This kind of lookup is useful in the case where several different
kinds of information are put in one column, e.g., a data source might be GenBank, PubMed,
AddGene, etc. Additionally, it allows the formatting of a URL rather than being a direct
remapping in the way a simple sheet look up is.
The final kind of lookup is Object ID lookup. The Object ID Lookup means that the
9
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint

Figure 7: Replacement Lookup Sheet. This is an example of a replacement lookup sheet
referenced by the column definitions sheet. In this case the prefix found in column B is
used to identify the correct URL in column and the suffix is placed in the URL at the
location indicated by {REPLACE HERE}. So for the case PMID:24295448, the PMID key
gives the string https://pubmed.ncbi.nlm.nih.gov/{REPLACE_HERE}, which is changed to
https://pubmed.ncbi.nlm.nih.gov/24295448.
cell value is converted to a URI. For example, a part called GFP promoter can be referenced
using that name, but this reference can then be converted to the URI http://www.examples.
org/gfp_promoter using this lookup. This type of lookup only works for SBOL objects.
If the parent lookup column is also true, then it indicates that the current object is added
as a property to the referenced object rather than vice versa. For example, in a row for a
component definition called pAOX1 (Figure 3), there may be a collection column with the
collection name: Promoter Collection. If the Parent Lookup is true, then rather than adding
Promoter Collection to pAOX1 using the sbol member attribute, the pAOX1 URI is added
to the Promoter Collection object using the sbol member attribute.
Excel-to-SBOL Conversion Algorithm
The initialization and column definition sheets are used by the Excel-to-SBOL conversion
algorithm shown in Algorithm 1. The full code for the Excel-to-SBOL converter can be found
at https://github.com/SynBioDex/Excel-to-SBOL. This algorithm proceeds as follows.
First, it reads in the initialization, column definition, and data sheets. It then adds any
additional properties specified on the Init sheet to the column definitions sheet as well as the
appropriate data sheet. Next, it creates an SBOL Document, and it begins parsing all the
sheets that have been marked in the initialization sheet as containing SBOL content. To do
this, it identifies the displayId and SBOL object type columns, and it creates an SBOL object
10
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint

of the specified type for each entry found with a URI constructed from the displayId found.
Finally, it parses all the other columns in each sheet as specified in the column definitions
sheet. First, it splits the entries found using the Split On field. Next, it converts the value
found, if needed, using the lookup method specified in the column definition sheet for that
entry. Then, it checks the converted cell value against the regular expression provided in
the Pattern field. Finally, it adds the property and value to the corresponding SBOL object
using the specified SBOL Term and Namespace URL.
Algorithm 1: Excel-to-SBOL
Input: Filled Excel Template
Output: SBOL File
Initialization
Read in Init Sheet and create a list of sheets with SBOL objects in them
Read in SBOL Version to Convert to based on Init Sheet
Read in all Sheets based on Init Sheet
Read in Column Definitions Sheet
Add any columns defined on the Init Sheet to the appropriate sheets and add
definitions for them in the Column Definitions Sheet
Parse Objects
Create SBOL Document
for sheet in SBOL Object Sheets do
Find Display ID Column and SBOL Object Type Column
for SBOL Object in sheet do
Create SBOL Object
Save SBOL Object in a dictionary common name:URI
Parse Columns
for sheet in SBOL Object Sheets do
for Row in sheet do
for Column in Row do
Split the cell value based on Column Definitions Sheet split column
Convert cell value using Ontology Lookups found in Column Definitions
Sheet
Check that the converted cell value match the pattern specified in the
Column Definitions Sheet
Convert the cell value based on SBOL Term, Namespace URL and
Parental Lookup values from the Column Definitions Sheet
11
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint
SBOL-to-Excel
The SBOL-to-Excel converter performs the opposite task of taking SBOL data and con-
verting it into an Excel spreadsheet. This converter reads an SBOL file in any standard
RDF format (e.g., XML, JSON-LD, Turdle, N-triples), and for every SBOL object found,
it creates a row in a spreadsheet that has a column for each property of the object. In this
case, the RDFlib python library is used rather than the more specialized SBOL libraries in
order to allow the conversion of different SBOL object types in any version of SBOL whilst
maintaining code simplicity.
An example Excel spreadsheet generated by the SBOL-to-Excel converter from the SBOL
generated for the library example presented earlier is shown in Figure 8. This spreadsheet
looks similar to the templates described earlier, but has a few key differences. First, all
column names are based on the property name rather than human readable column names.
Second, no Init or Column Definitions sheet is present in the output. Finally, many of the
properties are full URLs rather than human readable names. Future work will address these
differences to allow full round-tripping of data from Excel to SBOL and back again.
The algorithm used by the SBOL-to-Excel Converter is shown in Algorithm 2. The full
code for the SBOL-to-Excel Converter can be found at https://github.com/SynBioDex/
SBOL-to-Excel.
The algorithm first loads SBOL data into a dictionary with a dictionary of predicates
containing a list of objects for each subject. This is then converted to a pandas DataFrame.
Next, the predicate prefixes are converted to more human readable forms (e.g. “http:
//purl.org/dc/terms” to ‘dcterms’). Then the columns are reordered according to a hard-
coded priority system and any columns in a hard-coded removal list are removed. Finally,
each SBOL object type is output to its own sheet.
12
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint

(a) Collection Sheet
(b) ComponentDefinition Sheet
(c) Sequence Sheet
Figure 8: Spreadsheet output by the SBOL-to-Excel Converter. This is an example output
for the SBOL example shown in the Excel-to-SBOL section. Note that each SBOL object
type is split into separate sheets.
13
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint

Algorithm 2: SBOL to Excel
Input: SBOL File
Output: Excel Filled Template
Initialization
Load SBOL document
Initialize output path
Parse Graph Subjects
Initialize Data Frame
for Subject, Predicate, Object in SBOL document do
Add items to Data Frame
Process Column Names
for Column Name in Predicates
Process the property URLs into human readable values to be set as the
column names
return processed Column Name
Reorder and Drop Columns
Any Data Frame columns found in the column list are reordered according to the
order in the column list and are moved to the front. The rest of the columns
maintain the same order.
Get Data Frame with list of new columns
Drop unnecessary columns from Data Frame.
return Data Frame with list of dropped columns
Dataframe to Excel Processing
for SBOL Object Type in Data Frame
Create a new Work Sheet
Add all cell values to the Work Sheet
14
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint

SynBioHub Plugins
SynBioHub plugins for the Excel-SBOL Converter were developed to enable the converter
to be integrated into a data processing workflow using the SynBioHub repository (see Fig-
ure 9). SynBioHub plugins are a way to create modular extensions to the capabilities of the
SynBioHub repository.
44
The plugins developed as part of this project allow the integration
of SynBioHub into synthetic biology workflows with spreadsheets. A submit plugin was de-
veloped for the Excel-to-SBOL converter. This plugin takes in Excel spreadsheet templates,
converts them to SBOL, and uploads the converted SBOL to SynBioHub. Similarly, a down-
load plugin was developed using the SBOL-to-Excel library that allows a user to download
SBOL stored in SynBioHub in the form of an Excel spreadsheet.
Figure 9: Integration of the Excel-SBOL Converter with SynBioHub via plugins. Submit:
When a user uploads an Excel spreadsheet template to the submit endpoint, SynBioHub
sends it to the Excel-to-SBOL plugin, which returns SBOL to be deposited in SynBioHub.
Download: If a user requests an Excel spreadsheet template to be downloaded from Syn-
BioHub, SynBioHub sends the appropriate SBOL to the SBOL-to-Excel plugin. The plugin
returns an Excel spreadsheet to SynBioHub, which is then returned to the user.
15
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint
Case Studies
We now illustrate the operation of the Excel-SBOL Converter with two case studies, one
focused on the upload of experimental data, the other focused on downloading a part library.
Experimental Data Sheets
Capturing experimental data with Excel-to-SBOL allows researchers to more easily associate
measurement metadata with genetic part information. The experimental metadata may in-
clude descriptions of experimental samples, study conditions, assay types, and equipment.
All this information can be captured in the SBOL data standard. To process experimen-
tal data, an Excel spreadsheet template was created that includes sheets for Studies (i.e.,
experimental replicates), Assays (i.e., experiments), Sample Designs (i.e., the media, strain,
and vector used), Samples (i.e., an individually measured instance of a sample design), and
Measurements (i.e., the data collected).
In the example, shown in Figure 10, each experimental study is conducted over a period
of twenty-five hours with measurements taken every 15 minutes. This experimental study
is repeated two times, with the study composed of two assays and each assay comprising of
two samples with two sample designs, for a total of four samples. Each of these four samples
then has two signals (i.e., optical density (OD), green fluorescent protein (GFP)) measured
for 100 time points, resulting in a total of 800 measurements.
The information in this spreadsheet can be uploaded for storage in the SynBioHub repos-
itory
45
after processing by the Excel-to-SBOL converter. The Excel-to-SBOL converter con-
verts each study into an SBOL Collection object, each assay into an SBOL Experiment
object, each sample design into an SBOL Module Definition object, and each sample into an
SBOL ExperimentalData object. However, Measurements are not converted to SBOL and
are not uploaded to SynBioHub. In the future, we plan to upload the measurement data to
the Flapjack repository
46
(a repository for the storage of experimental measurement data).
16
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint

Figure 10: Excel-to-SBOL example for a collection of experiments. Top: Diagram showing
the design of an example experiment comprising a study with measurements every 15 minutes
over a period of twenty-five hours with two repeats. For each repeat, there is one plate per
repeat with one assay per plate. Each plate has two samples per assay for a total of four
samples at two different sample design setups. Measuring each sample at each of the 100
time points results in 800 data points. Bottom: Given this experimental setup, five different
sheets are used to describe and connect information for Studies, Assays, Sample Designs,
Samples, and Measurements. Note the way the sheets are linked via IDs. For example, assays
identify the study they are part of using the “Study ID” column and similarly, samples to
assay with “Assay ID”, and measurements to sample with “Sample ID”.
17
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint
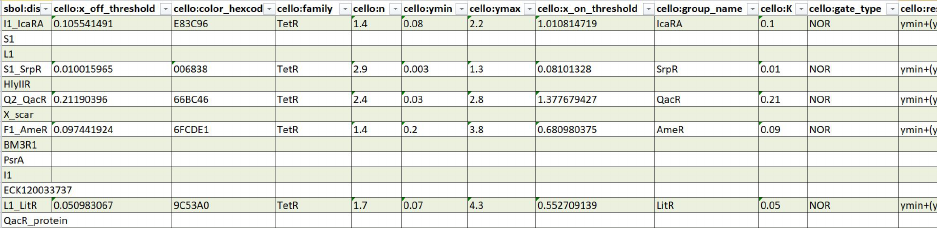
Cello
The Cello library
47
was chosen as a case study for the SBOL-to-Excel Converter due the
variety of SBOL objects that it contains, including objects with a diverse set of custom
annotations. In order to achieve compliance with these varied SBOL types, it is key that
the SBOL-to-Excel Converter has a way of dynamically manipulating these types. With
the XML parsing capabilities offered by RDFLib, the SBOL-to-Excel Converter is able to
successfully process and organize all of the data into a 15 sheets in an Excel document. The
conversion to Excel facilitates the analysis of the SBOL document for a user that prefers
spreadsheets. The results of the Cello conversion can be seen in Figure 11.
Figure 11: Example of a part of a sheet (ComponentDefinition) output by the SBOL-to-
Excel Converter from the conversion of the Cello part Library.
47
In total 15 sheets are output
for: Range, Collection, Interaction, ComponentDefinition, ModuleDefinition, Participation,
FunctionalComponent, Sequence, Attachment, SequenceAnnotation, Component, Activity,
Agent, Association, and Usage. Note the human readable column names.
Discussion
We have presented two Python converter libraries: Excel-to-SBOL and SBOL-to-Excel. The
development of these libraries simplifies the incorporation of SBOL into existing synthetic
biology workflows that make use of spreadsheets for data storage and exchange. The Excel-
to-SBOL converter allows multiple sheets with different SBOL object types to be processed.
The converter can be used to convert spreadsheet columns into any RDF properties, and
can output either SBOL Version 2 or 3 documents, as well as validating information using
18
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint
user supplied regular expressions and converting user-friendly names into ontology terms.
Its complement, the SBOL-to-Excel Converter, can process any RDF file and turn it into
a spreadsheet. This allows both SBOL2 and SBOL3 documents to be converted to spread-
sheets. The conversion makes the values more human readable, and splits different SBOL
object types across sheets.
While the Excel-SBOL Converter is already useful for many applications, there are several
improvements planned for the future. For the Excel-to-SBOL Converter, real-time sheet
checking using Excel plugins would enable errors to be detected and fixed more efficiently.
Second, more standard Excel spreadsheet templates with example data should be created
to provide a starting point for both users and template creators. For the SBOL-to-Excel
Converter, the next step is to record the conversion process into a column definition sheet
in order to support round-trip conversion back to SBOL.
Finally, the case studies presented here both indicate that more thought needs to be
given to development of core standards regarding the information to be collected and shared
about experiments and parts. There is currently no consensus regarding what information
is desirable to share or how to organize it. Excel templates can be made available using
the Excel-SBOL Converter, however, it may be possible to help in developing a de facto
standard for information to be collected, and such a minimum information standard should
be further considered.
Methods
Below is a more in depth description of the methods used by the Excel-to-SBOL and SBOL-
to-Excel Converters, as well as the methods used in the case studies. Note that all code was
written in Python.
19
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint
Excel-to-SBOL
This converter relies on a set of Python modules to function. The modules used and their
functionality in the converter is explained below.
• Openpyxl is a Python library to read and write Excel files. This library is used
together with Pandas to read in the data from the Excel workbook and store it as a
data frame.
• pandas is a data analysis library. It is used for the reading in of the Excel workbook
and the further storage and manipulation of the data.
• pySBOL2 and pySBOL3 are libraries for the reading, writing, and manipulation of
SBOL data. These libraries are used to create the SBOL objects required and write
them out to an SBOL document.
• Tyto is a tool to make the semantic web more accessible. It is used to convert cell
values to ontology terms.
SBOL-to-Excel
SBOL-to-Excel relies on a set of libraries to ensure the smooth processing of a given SBOL
document. These libraries facilitate advanced processing procedures, and ensure that the
data is properly output to Excel for the user to analyze.
• RDFLib is a python package that enables the user to work with the Resource De-
scription Framework (RDF). RDF serves as an important proponent to this project.
Its functionality includes the ability to parse through RDF triples within an Excel file.
With access to the subject, object, and predicate triples, the library made it possible
to extract these values, and convert them into a form that can be easily interpreted in
order to eventually be output to Excel.
20
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint
• Openpyxl is a Python library to read and write Excel files. The Openpyxl library is
crucial to the Converter, as its functionality allows the writing of SBOL data into the
Excel spreadsheet. It is then also used to format the spreadsheet (including creating
the hyperlinks).
• Pandas is a python library that provides data structures with capabilities for the
accessing and manipulating of data that it holds, and does so with the information
processed from the SBOL collection. This occurs specifically when the data is being
read in. The subjects, objects, and predicates are set into the pandas data frame in
order to be processed by various modules, before being output into Excel.
• Validators is used to check items expected to be URLs are valid URLS.
• Pytest makes it easy to write small tests, as well as, scales to support testing for large
applications. This library enabled modular testing, ensuring that the correct forms of
data were being passed through the converter at the appropriate points.
Acknowledgement
JA, JM, and CM are supported by the National Science Foundation under Grant No.
1939892. JM is additionally supported by a Dean’s Graduate Assistantship at the Univer-
sity of Colorado Boulder. JA is additionally supported by a CU Boulder Discovery Learning
Apprenticeship. IP was supported by the Google Summer of Code and the Bisch¨ofliche Stu-
dienf¨orderung Cusanuswerk. PS was supported by the SBOL Industrial Consortium. JB and
CM are partially supported by Air Force Research Laboratory (AFRL) contracts FA8750-
17-C-0184 and FA8750-17-C-0229. SynBioHub and the Excel-SBOL Converter plugins are
run on a Microsoft Azure Server provided by Microsoft Research. This document does not
contain technology or technical data controlled under either U.S. International Traffic in
Arms Regulation or U.S. Export Administration Regulations. Any opinions, findings, and
21
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint
conclusions or recommendations expressed in this material are those of the author(s) and
do not necessarily reflect the views of the funding agencies. All authors contributed to the
writing of this manuscript.
Author Contributions
All authors contributed to the writing of this manuscript. JM worked on Excel-to-SBOL,
supervising IP in the initial design and then taking over. JA worked on SBOL-to-Excel
with the help of JM. SS created the experimental data case study. PS is working on Tyto
integration into Excel. CM supervised the project. JB contributed guidance and the initial
DARPA spreadsheets.
Conflicts of Interest
The authors declare no conflicts of interest.
Supporting Information Available
Excel2SBOL
• GitHub: https://github.com/SynBioDex/Excel-to-SBOL
• Documentation: https://github.com/SynBioDex/Excel-to-SBOL/wiki
• Plugin: https://github.com/SynBioHub/Plugin-Submit-Excel2SBOL
• PyPI: https://pypi.org/project/excel2sbol/
SBOL2Excel
• GitHub: https://github.com/SynBioDex/SBOL-to-Excel
• Documentation: https://github.com/SynBioDex/SBOL-to-Excel/wiki
• Plugin: https://github.com/SynBioHub/Plugin-Download-SBOL2Excel
• PyPI: https://pypi.org/project/sbol2excel/
22
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint

Suplemental Filesl
• Template for Excel to SBOL Conversion SBOL2 simple parts template.xlsx
• Output of SBOL to Excel sbol2excel output.xlsx
• Case Study Excel to SBOL (Experimental Data) flapjack compiler sbol3 v023.xlsx
• Case Study SBOL to Excel (Cello) cello.xlsx
References
(1) Khalil, A. S.; Collins, J. J. Synthetic biology: applications come of age. Nature Reviews
Genetics 2010, 11, 367–379.
(2) Hucka, M.; Finney, A.; Sauro, H. M.; Bolouri, H.; Doyle, J. C.; Kitano, H.; Arkin, A. P.;
Bornstein, B. J.; Bray, D.; Cornish-Bowden, A. et al. The systems biology markup
language (SBML): a medium for representation and exchange of biochemical network
models. Bioinformatics (Oxford, England) 2003, 19, 524–531.
(3) Hucka, M.; Nickerson, D. P.; Bader, G. D.; Bergmann, F. T.; Cooper, J.; Demir, E.;
Garny, A.; Golebiewski, M.; Myers, C. J.; Schreiber, F. et al. Promoting Coordinated
Development of Community-Based Information Standards for Modeling in Biology:
The COMBINE Initiative. Frontiers in Bioengineering and Biotechnology 2015, 3, 19.
(4) Waltemath, D.; Adams, R.; Bergmann, F. T.; Hucka, M.; Kolpakov, F.; Miller, A. K.;
Moraru, I. I.; Nickerson, D.; Sahle, S.; Snoep, J. L. et al. Reproducible computational
biology experiments with SED-ML–the Simulation Experiment Description Markup
Language. BMC systems biology 2011, 5, 198.
(5) Nov`ere, N. L.; Hucka, M.; Mi, H.; Moodie, S.; Schreiber, F.; Sorokin, A.; Demir, E.;
Wegner, K.; Aladjem, M. I.; Wimalaratne, S. M. et al. The Systems Biology Graphical
Notation. Nature Biotechnology 2009, 27, 735–741.
23
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint
(6) Galdzicki, M.; Clancy, K. P.; Oberortner, E.; Pocock, M.; Quinn, J. Y.; Ro-
driguez, C. A.; Roehner, N.; Wilson, M. L.; Adam, L.; Anderson, J. C. et al. The
Synthetic Biology Open Language (SBOL) provides a community standard for commu-
nicating designs in synthetic biology. Nature Biotechnology 2014, 32, 545–550.
(7) Gleeson, P.; Crook, S.; Cannon, R. C.; Hines, M. L.; Billings, G. O.; Farinella, M.;
Morse, T. M.; Davison, A. P.; Ray, S.; Bhalla, U. S. et al. NeuroML: A Language
for Describing Data Driven Models of Neurons and Networks with a High Degree of
Biological Detail. PLOS Computational Biology 2010, 6, e1000815.
(8) Cannon, R. C.; Gleeson, P.; Crook, S.; Ganapathy, G.; Marin, B.; Piasini, E.; Sil-
ver, R. A. LEMS: a language for expressing complex biological models in concise and
hierarchical form and its use in underpinning NeuroML 2. Frontiers in Neuroinformatics
2014, 8, 79.
(9) Faeder, J. R.; Blinov, M. L.; Hlavacek, W. S. In Systems Biology; Maly, I. V., Ed.;
Methods in Molecular Biology; Humana Press, 2009; Vol. 500; p 113–167.
(10) Bergmann, F. T.; Adams, R.; Moodie, S.; Cooper, J.; Glont, M.; Golebiewski, M.;
Hucka, M.; Laibe, C.; Miller, A. K.; Nickerson, D. P. et al. COMBINE archive and
OMEX format: one file to share all information to reproduce a modeling project. BMC
bioinformatics 2014, 15, 369.
(11) Gennari, J. H.; Neal, M. L.; Galdzicki, M.; Cook, D. L. Multiple ontologies in action:
composite annotations for biosimulation models. Journal of Biomedical Informatics
2011, 44, 146–154.
(12) Wolstencroft, K.; Krebs, O.; Snoep, J. L.; Stanford, N. J.; Bacall, F.; Golebiewski, M.;
Kuzyakiv, R.; Nguyen, Q.; Owen, S.; Soiland-Reyes, S. et al. FAIRDOMHub: a reposi-
tory and collaboration environment for sharing systems biology research. Nucleic Acids
Research 2017, 45, D404–D407.
24
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint
(13) Wittig, U.; Kania, R.; Golebiewski, M.; Rey, M.; Shi, L.; Jong, L.; Algaa, E.; Wei-
demann, A.; Sauer-Danzwith, H.; Mir, S. et al. SABIO-RK–database for biochemical
reaction kinetics. Nucleic Acids Research 2012, 40, D790–796.
(14) Nickerson, D.; Atalag, K.; de Bono, B.; Geiger, J.; Goble, C.; Hollmann, S.; Lonien, J.;
M¨uller, W.; Regierer, B.; Stanford, N. J. et al. The Human Physiome: how standards,
software and innovative service infrastructures are providing the building blocks to
make it achievable. Interface Focus 2016, 6, 20150103.
(15) Yu, T.; Lloyd, C. M.; Nickerson, D. P.; Cooling, M. T.; Miller, A. K.; Garny, A.;
Terkildsen, J. R.; Lawson, J.; Britten, R. D.; Hunter, P. J. et al. The Physiome Model
Repository 2. Bioinformatics (Oxford, England) 2011, 27, 743–744.
(16) Wolstencroft, K.; Owen, S.; Horridge, M.; Jupp, S.; Krebs, O.; Snoep, J.; Du Preez, F.;
Mueller, W.; Stevens, R.; Goble, C. Stealthy annotation of experimental biology by
spreadsheets. Concurrency and Computation: Practice and Experience 2013, 25, 467–
480.
(17) Watanabe, L.; Nguyen, T.; Zhang, M.; Zundel, Z.; Zhang, Z.; Madsen, C.; Roehner, N.;
Myers, C. IBIOSIM 3: A Tool for Model-Based Genetic Circuit Design. ACS Synthetic
Biology 2019, 8, 1560–1563.
(18) Myers, C. J.; Barker, N.; Jones, K.; Kuwahara, H.; Madsen, C.; Nguyen, N.-P. D.
iBioSim: A Tool for the Analysis and Design of Genetic Circuits. Bioinformatics 2009,
25, 2848–2849.
(19) Roehner, N.; Myers, C. J. Directed Acyclic Graph-Based Technology Mapping of Ge-
netic Circuit Models. ACS Synthetic Biology 2014, 3, 543–555.
(20) Chen, Y.; Zhang, S.; Young, E. M.; Jones, T. S.; Densmore, D.; Voigt, C. A. Genetic
circuit design automation for yeast. 5, 1349–1360.
25
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint

(21) Nielsen, A. A. K.; Der, B. S.; Shin, J.; Vaidyanathan, P.; Paralanov, V.; Strychal-
ski, E. A.; Ross, D.; Densmore, D.; Voigt, C. A. Genetic circuit design automa-
tion. 352, Publisher: American Association for the Advancement of Science eprint:
https://science.sciencemag.org/content/352/6281/aac7341.full.pdf.
(22) Baig, H.; Madsen, J. A Top-down Approach to Genetic Circuit Synthesis and Opti-
mized Technology Mapping. 9th International Workshop on Bio-Design Automation
(Pittsburgh, PA). 2017; pp 1–2.
(23) Gaspar, P.; Oliveira, J. L.; Frommlet, J.; Santos, M. A. S.; Moura, G. EuGene: Max-
imizing Synthetic Gene Design for Heterologous Expression. Bioinformatics 2012, 28,
2683–2684.
(24) Chen, J.; Densmore, D.; Ham, T. S.; Keasling, J. D.; Hillson, N. J. DeviceEditor Visual
Biological CAD Canvas. Journal of Biological Engineering 2012, 6, 1.
(25) Terry, L.; Earl, J.; Thayer, S.; Bridge, S.; Myers, C. J. SBOLCanvas: A Visual Editor
for Genetic Designs. ACS Synthetic Biology 2021, 10, 1792–1796.
(26) Czar, M. J.; Cai, Y.; Peccoud, J. Writing DNA with GenoCADTM. Nucleic Acids
Research 2009, 37, W40–W47.
(27) McLaughlin, J. A.; Myers, C. J.; Zundel, Z.; Mısırlı, G.; Zhang, M.; Ofiteru, I. D.;
Go˜ni-Moreno, A.; Wipat, A. SynBioHub: A Standards-Enabled Design Repository for
Synthetic Biology. ACS Synthetic Biology 2018, 7, 682–688.
(28) Roehner, N.; Beal, J.; Clancy, K.; Bartley, B.; Misirli, G.; Gr¨unberg, R.; Oberortner, E.;
Pocock, M.; Bissell, M.; Madsen, C. et al. Sharing Structure and Function in Biological
Design with SBOL 2.0. ACS Synthetic Biology 2016, 5, 498–506.
(29) McLaughlin, J. A.; Beal, J.; Mısırlı, G.; Gr¨unberg, R.; Bartley, B. A.; Scott-Brown, J.;
Vaidyanathan, P.; Fontanarrosa, P.; Oberortner, E.; Wipat, A. et al. The Synthetic Bi-
26
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint
ology Open Language (SBOL) Version 3: Simplified Data Exchange for Bioengineering.
Frontiers in Bioengineering and Biotechnology 2020, 8 .
(30) Maccagnan, A.; Riva, M.; Feltrin, E.; Simionati, B.; Vardanega, T.; Valle, G.; Can-
nata, N. Combining ontologies and workflows to design formal protocols for biological
laboratories. Automated Experimentation 2010, 2, 3.
(31) Curty, R. G.; Crowston, K.; Specht, A.; Grant, B. W.; Dalton, E. D. Attitudes and
norms affecting scientists’ data reuse. PLOS ONE 2017, 12, e0189288.
(32) Pasquetto, I. V.; Borgman, C. L.; Wofford, M. F. Uses and Reuses of Scientific Data:
The Data Creators’ Advantage. Harvard Data Science Review 2019, 1 .
(33) Zuiderwijk, A.; Shinde, R.; Jeng, W. What drives and inhibits researchers to share and
use open research data? A systematic literature review to analyze factors influencing
open research data adoption. PLOS ONE 2020, 15, e0239283.
(34) Costello, M. J.; Michener, W. K.; Gahegan, M.; Zhang, Z.-Q.; Bourne, P. E. Biodiversity
data should be published, cited, and peer reviewed. Trends in Ecology & Evolution
2013, 28, 454–461.
(35) Borgman, C. L. The conundrum of sharing research data. Journal of the American
Society for Information Science and Technology 2012, 63, 1059–1078.
(36) Frey, K.; Hafner, A.; Pucker, B. The Reuse of Public Datasets in the Life Sciences:
Potential Risks and Rewards; 2020.
(37) Booth, I. R. SysMO: back to the future. Nature Reviews Microbiology 2007, 5, 566–566.
(38) Spellman, P. T.; Miller, M.; Stewart, J.; Troup, C.; Sarkans, U.; Chervitz, S.; Bern-
hart, D.; Sherlock, G.; Ball, C.; Lepage, M. et al. Design and implementation of mi-
croarray gene expression markup language (MAGE-ML). Genome Biology 2002, 3,
research0046.1.
27
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint
(39) Wolstencroft, K.; Owen, S.; Horridge, M.; Krebs, O.; Mueller, W.; Snoep, J. L.;
du Preez, F.; Goble, C. RightField: embedding ontology annotation in spreadsheets.
Bioinformatics 2011, 27, 2021–2022.
(40) Urquiza-Garc´ıa, U.; Zieli´nski, T.; Millar, A. J. Better research by efficient sharing:
evaluation of free management platforms for synthetic biology designs. Synthetic Biology
2019, 4, ysz016.
(41) Zieli´nski, T.; Hay, J.; Romanowski, A.; Nenninger, A.; McCormick, A.; Millar, A. J.
SynBio2Easy—a biologist-friendly tool for batch operations on SBOL designs with Ex-
cel inputs. Synthetic Biology 2022, 7, ysac002.
(42) Bartley, B. A. Tyto: A Python Tool Enabling Better Annotation Practices for Synthetic
Biology Data-Sharing. ACS Synthetic Biology 2022,
(43) Eilbeck, K.; Lewis, S. E.; Mungall, C. J.; Yandell, M.; Stein, L.; Durbin, R.; Ash-
burner, M. The Sequence Ontology: a tool for the unification of genome annotations.
Genome Biology 2005, 6, R44.
(44) Mante, J.; Zundel, Z.; Myers, C. Extending SynBioHub’s Functionality with Plugins.
ACS Synthetic Biology 2020, 9, 1216–1220.
(45) McLaughlin, J. A.; Myers, C. J.; Zundel, Z.; Mısırlı, G.; Zhang, M.; Ofiteru, I. D.;
Goni-Moreno, A.; Wipat, A. SynBioHub: a standards-enabled design repository for
synthetic biology. ACS synthetic biology 2018, 7, 682–688.
(46) Yanez Feliu, G.; Earle Gomez, B.; Codoceo Berrocal, V.; Mu˜noz Silva, M.; Nu˜n˜ez, I. N.;
Matute, T. F.; Arce Medina, A.; Vidal, G.; Vidal C´e´sspedes, C.; Dahlin, J. et al.
Flapjack: Data management and analysis for genetic circuit characterization. ACS
Synthetic Biology 2020, 10, 183–191.
28
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint
(47) Jones, T. S.; Oliveira, S.; Myers, C. J.; Voigt, C. A.; Densmore, D. Genetic circuit
design automation with Cello 2.0. Nature Protocols 2022, 1–17.
29
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint

Graphical TOC Entry
30
.CC-BY-NC-ND 4.0 International licenseavailable under a
(which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
The copyright holder for this preprintthis version posted August 31, 2022. ; https://doi.org/10.1101/2022.08.31.505873doi: bioRxiv preprint
